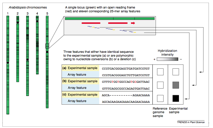
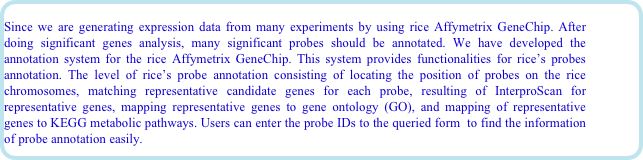
After the completion of the rice genome sequencing project, Rice Gene Discovery Unit aim to use the bioinformatics software, and database for discovery rice genes, which responsible for the economic trait such as the submergence tolerance, salt tolerance, disease resistances, and so on. To discover those important rice genes, RiceGeneThresher, a public resource for the rice genomic and bioinformatics software and platform, has been developed. RiceGeneThresher funded by the National Center for Genetic Engineering and Biotechnology (BIOTEC), Thailand. The objective of developing RiceGeneThresher is to provide rice researchers as well as breeders an expert and database management system for gene discovery by providing full access to integrated multiplatform relational database of heterogeneous rice genome resource for decision-making and candidate gene selection. Since the post-genome era up coming, we are generating expression data from many experiments by using rice Affymetrix GeneChip (www.affymetrix.com). RiceGeneThresher also aim to provide analysis tool for analyzing microarray analysis by using the Bioconductor set of software. Recently, we also released the tool for discovery the single-feature polymorphism analysis (SFP) by using rice Affymetrix GeneChip.







About Project
Current Project Tools and Software :
Rice Gene Discovery Unit. Kasetsart University Kamphaeng Saen Campus
Kamphaeng Saen, Nakhon Pathom 73140 Thailand
Tel. (66) 2942 8200 Ext.3688-91 Ext.103, (66) 3435 5192-4 Fax (66) 3435 5196
Information Assistant E-mail:supat.thongjuea@gmail.com


Supported By :